publications
2025
- Neuron
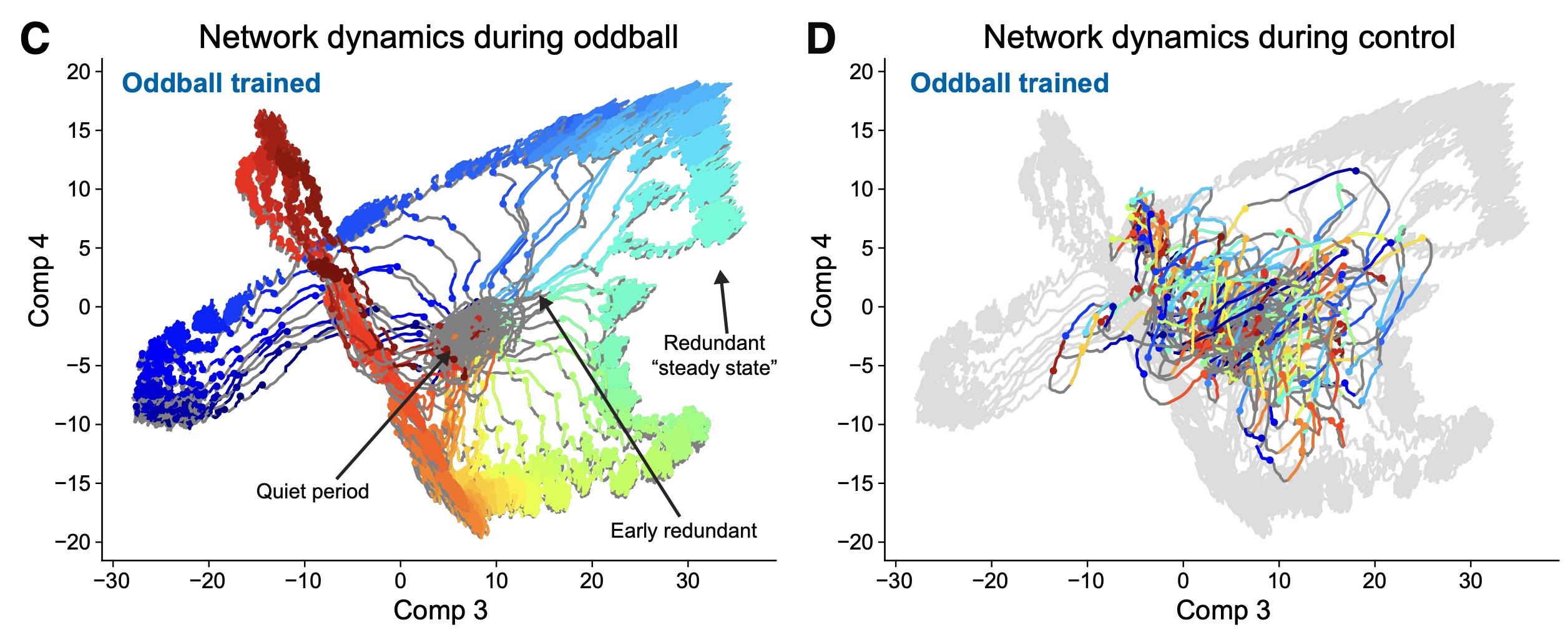 Slow cortical dynamics generate context processing and novelty detectionYuriy Shymkiv, Jordan P Hamm, Sean Escola, and 1 more authorNeuron, 2025
Slow cortical dynamics generate context processing and novelty detectionYuriy Shymkiv, Jordan P Hamm, Sean Escola, and 1 more authorNeuron, 2025The cortex amplifies responses to novel stimuli while suppressing redundant ones. Novelty detection is necessary to efficiently process sensory information and build predictive models of the environment, and it is also altered in schizophrenia. To investigate the circuit mechanisms underlying novelty detection, we used an auditory “oddball” paradigm and two-photon calcium imaging to measure responses to simple and complex stimuli across mouse auditory cortex. Stimulus statistics and complexity generated specific responses across auditory areas. Neuronal ensembles reliably encoded auditory features and temporal context. Interestingly, stimulus-evoked population responses were particularly long lasting, reflecting stimulus history and affecting future responses. These slow cortical dynamics encoded stimulus temporal context and generated stronger responses to novel stimuli. Recurrent neural network models trained on the oddball task also exhibited slow network dynamics and recapitulated the biological data. We conclude that the slow dynamics of recurrent cortical networks underlie processing and novelty detection.
@article{shymkiv2025slow, title = {Slow cortical dynamics generate context processing and novelty detection}, author = {Shymkiv, Yuriy and Hamm, Jordan P and Escola, Sean and Yuste, Rafael}, journal = {Neuron}, year = {2025}, publisher = {Elsevier}, doi = {https://doi.org/10.1016/j.neuron.2025.01.011}, }
2024
- Nat Neuro
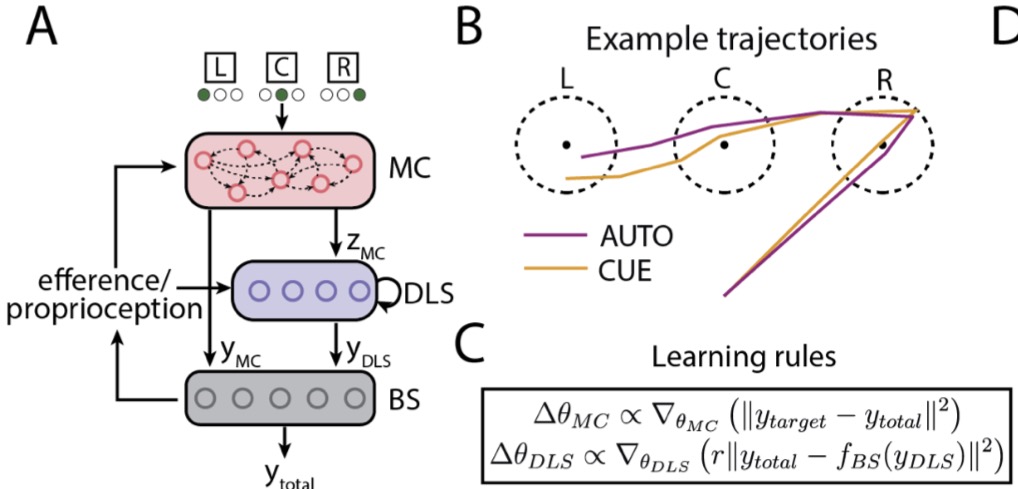 The role of motor cortex in motor sequence execution depends on demands for flexibilityKevin GC Mizes, Jack Lindsey, G Sean Escola, and 1 more authorNature Neuroscience, 2024
The role of motor cortex in motor sequence execution depends on demands for flexibilityKevin GC Mizes, Jack Lindsey, G Sean Escola, and 1 more authorNature Neuroscience, 2024The role of the motor cortex in executing motor sequences is widely debated, with studies supporting disparate views. Here we probe the degree to which the motor cortex’s engagement depends on task demands, specifically whether its role differs for highly practiced, or ‘automatic’, sequences versus flexible sequences informed by external cues. To test this, we trained rats to generate three-element motor sequences either by overtraining them on a single sequence or by having them follow instructive visual cues. Lesioning motor cortex showed that it is necessary for flexible cue-driven motor sequences but dispensable for single automatic behaviors trained in isolation. However, when an automatic motor sequence was practiced alongside the flexible task, it became motor cortex dependent, suggesting that an automatic motor sequence fails to consolidate subcortically when the same sequence is produced also in a flexible context. A simple neural network model recapitulated these results and offered a circuit-level explanation. Our results critically delineate the role of the motor cortex in motor sequence execution, describing the conditions under which it is engaged and the functions it fulfills, thus reconciling seemingly conflicting views about motor cortex’s role in motor sequence generation.
@article{mizes2024role, title = {The role of motor cortex in motor sequence execution depends on demands for flexibility}, author = {Mizes, Kevin GC and Lindsey, Jack and Escola, G Sean and {\"O}lveczky, Bence P}, journal = {Nature Neuroscience}, pages = {2466--2475}, year = {2024}, publisher = {Nature Publishing Group US New York}, doi = {https://doi.org/10.1038/s41593-024-01792-3}, volume = {27}, number = {12} } - Cell Reports
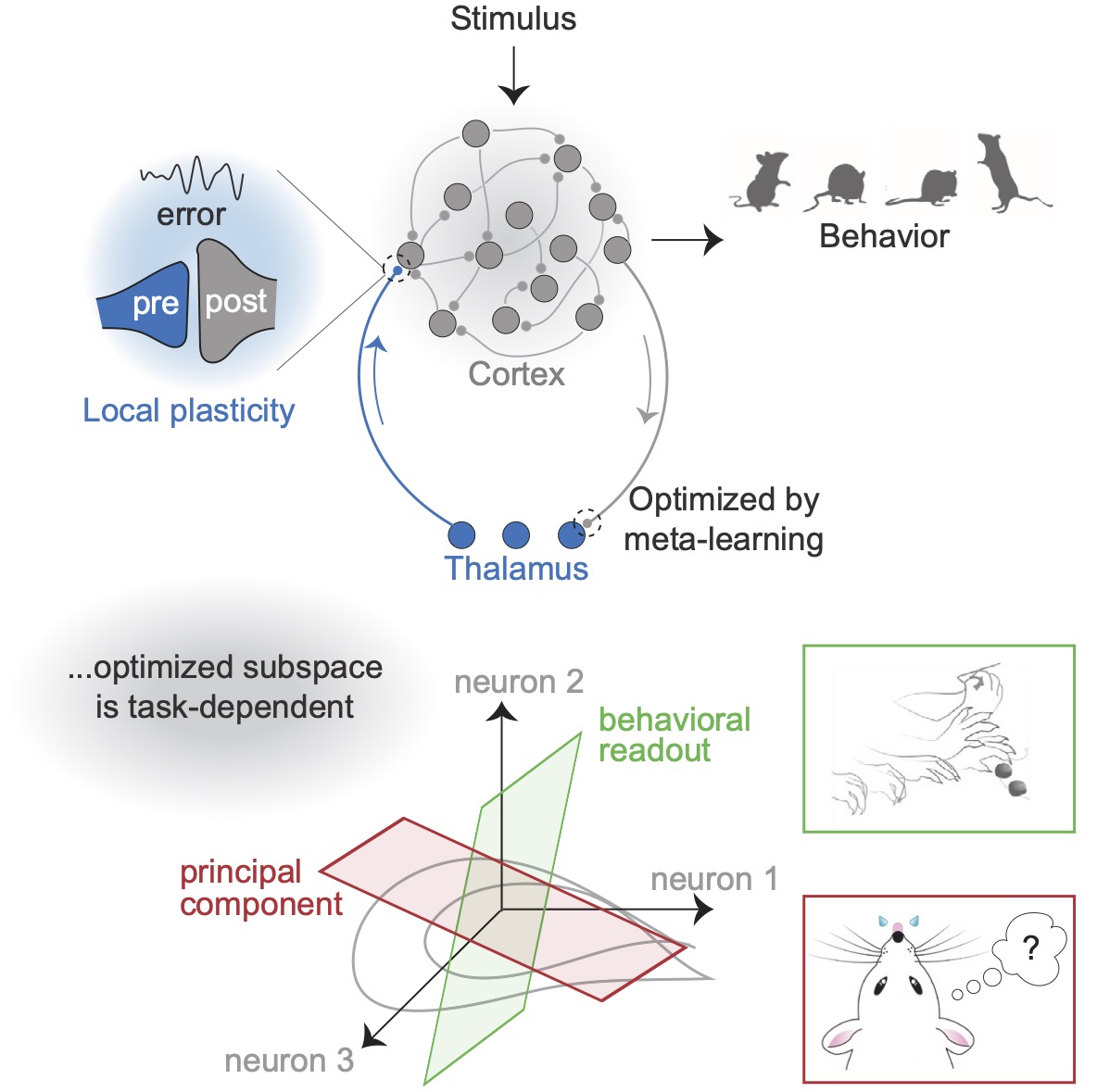 Specific connectivity optimizes learning in thalamocortical loopsKaushik J Lakshminarasimhan, Marjorie Xie, Jeremy D Cohen, and 4 more authorsCell Reports, 2024
Specific connectivity optimizes learning in thalamocortical loopsKaushik J Lakshminarasimhan, Marjorie Xie, Jeremy D Cohen, and 4 more authorsCell Reports, 2024Thalamocortical loops have a central role in cognition and motor control, but precisely how they contribute to these processes is unclear. Recent studies showing evidence of plasticity in thalamocortical synapses indicate a role for the thalamus in shaping cortical dynamics through learning. Since signals undergo a compression from the cortex to the thalamus, we hypothesized that the computational role of the thalamus depends critically on the structure of corticothalamic connectivity. To test this, we identified the optimal corticothalamic structure that promotes biologically plausible learning in thalamocortical synapses. We found that corticothalamic projections specialized to communicate an efference copy of the cortical output benefit motor control, while communicating the modes of highest variance is optimal for working memory tasks. We analyzed neural recordings from mice performing grasping and delayed discrimination tasks and found corticothalamic communication consistent with these predictions. These results suggest that the thalamus orchestrates cortical dynamics in a functionally precise manner through structured connectivity.
@article{lakshminarasimhan2024specific, doi = {10.1016/j.celrep.2024.114059}, title = {Specific connectivity optimizes learning in thalamocortical loops}, author = {Lakshminarasimhan, Kaushik J and Xie, Marjorie and Cohen, Jeremy D and Sauerbrei, Britton A and Hantman, Adam W and Litwin-Kumar, Ashok and Escola, Sean}, journal = {Cell Reports}, volume = {43}, number = {4}, year = {2024}, publisher = {Elsevier} }
2023
- Nat Neuro
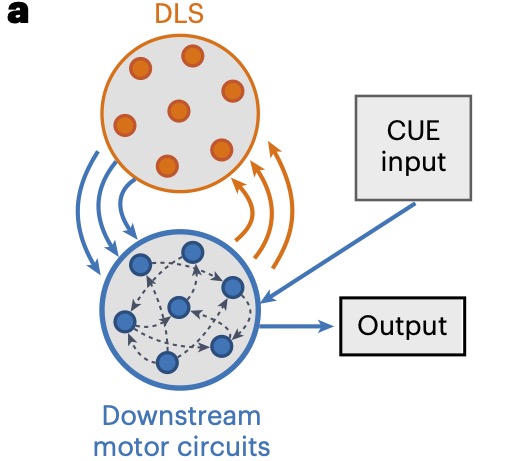 Dissociating the contributions of sensorimotor striatum to automatic and visually guided motor sequencesKevin GC Mizes, Jack Lindsey, G Sean Escola, and 1 more authorNature Neuroscience, 2023
Dissociating the contributions of sensorimotor striatum to automatic and visually guided motor sequencesKevin GC Mizes, Jack Lindsey, G Sean Escola, and 1 more authorNature Neuroscience, 2023The ability to sequence movements in response to new task demands enables rich and adaptive behavior. However, such flexibility is computationally costly and can result in halting performances. Practicing the same motor sequence repeatedly can render its execution precise, fast and effortless, that is, ‘automatic’. The basal ganglia are thought to underlie both types of sequence execution, yet whether and how their contributions differ is unclear. We parse this in rats trained to perform the same motor sequence instructed by cues and in a self-initiated overtrained, or ‘automatic,’ condition. Neural recordings in the sensorimotor striatum revealed a kinematic code independent of the execution mode. Although lesions reduced the movement speed and affected detailed kinematics similarly, they disrupted high-level sequence structure for automatic, but not visually guided, behaviors. These results suggest that the basal ganglia are essential for ‘automatic’ motor skills that are defined in terms of continuous kinematics, but can be dispensable for discrete motor sequences guided by sensory cues.
@article{mizes2023dissociating, doi = {10.1038/s41593-023-01431-3}, title = {Dissociating the contributions of sensorimotor striatum to automatic and visually guided motor sequences}, author = {Mizes, Kevin GC and Lindsey, Jack and Escola, G Sean and {\"O}lveczky, Bence P}, journal = {Nature Neuroscience}, volume = {26}, number = {10}, pages = {1791--1804}, year = {2023}, publisher = {Nature Publishing Group US New York} } - Nat Comm
 Catalyzing next-generation artificial intelligence through NeuroAIAnthony Zador, Sean Escola, Blake Richards, and 8 more authorsNature Communications, 2023
Catalyzing next-generation artificial intelligence through NeuroAIAnthony Zador, Sean Escola, Blake Richards, and 8 more authorsNature Communications, 2023Neuroscience has long been an essential driver of progress in artificial intelligence (AI). We propose that to accelerate progress in AI, we must invest in fundamental research in NeuroAI. A core component of this is the embodied Turing test, which challenges AI animal models to interact with the sensorimotor world at skill levels akin to their living counterparts. The embodied Turing test shifts the focus from those capabilities like game playing and language that are especially well-developed or uniquely human to those capabilities – inherited from over 500 million years of evolution – that are shared with all animals. Building models that can pass the embodied Turing test will provide a roadmap for the next generation of AI.
@article{zador2023catalyzing, doi = {10.1038/s41467-023-37180-x}, title = {Catalyzing next-generation artificial intelligence through NeuroAI}, author = {Zador, Anthony and Escola, Sean and Richards, Blake and {\"O}lveczky, Bence and Bengio, Yoshua and Boahen, Kwabena and Botvinick, Matthew and Chklovskii, Dmitri and Churchland, Anne and Clopath, Claudia and others}, journal = {Nature Communications}, volume = {14}, number = {1}, pages = {1597}, year = {2023}, publisher = {Nature Publishing Group UK London} }
2022
- Patterns
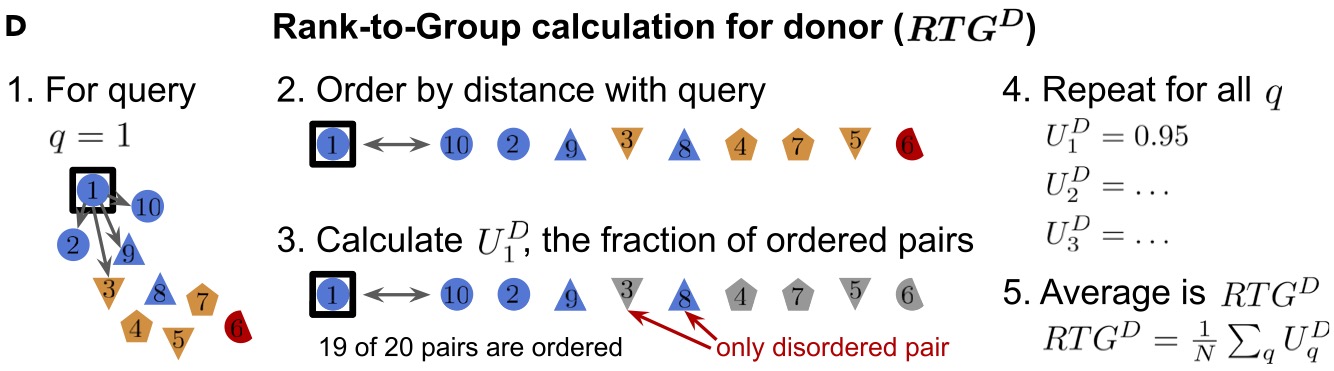 Hierarchical confounder discovery in the experiment-machine learning cycleAlex Rogozhnikov, Pavan Ramkumar, Rishi Bedi, and 2 more authorsPatterns, 2022
Hierarchical confounder discovery in the experiment-machine learning cycleAlex Rogozhnikov, Pavan Ramkumar, Rishi Bedi, and 2 more authorsPatterns, 2022The promise of machine learning (ML) to extract insights from high-dimensional datasets is tempered by confounding variables. It behooves scientists to determine if a model has extracted the desired information or instead fallen prey to bias. Due to features of natural phenomena and experimental design constraints, bioscience datasets are often organized in nested hierarchies that obfuscate the origins of confounding effects and render confounder amelioration methods ineffective. We propose a non-parametric statistical method called the rank-to-group (RTG) score that identifies hierarchical confounder effects in raw data and ML-derived embeddings. We show that RTG scores correctly assign the effects of hierarchical confounders when linear methods fail. In a public biomedical image dataset, we discover unreported effects of experimental design. We then use RTG scores to discover crossmodal correlated variability in a multi-phenotypic biological dataset. This approach should be generally useful in experiment-analysis cycles and to ensure confounder robustness in ML models.
@article{rogozhnikov2022hierarchical, doi = {10.1016/j.patter.2022.100451}, title = {Hierarchical confounder discovery in the experiment-machine learning cycle}, author = {Rogozhnikov, Alex and Ramkumar, Pavan and Bedi, Rishi and Kato, Saul and Escola, G Sean}, journal = {Patterns}, volume = {3}, number = {4}, year = {2022}, publisher = {Elsevier} } - JOSENeuromatch Academy: a 3-week, online summer school in computational neuroscienceBernard Marius Hart, Titipat Achakulvisut, Ayoade Adeyemi, and 8 more authorsJournal of Open Source Education, 2022
Neuromatch Academy (https://academy.neuromatch.io; (van Viegen et al., 2021)) was designed as an online summer school to cover the basics of computational neuroscience in three weeks. The materials cover dominant and emerging computational neuroscience tools, how they complement one another, and specifically focus on how they can help us to better understand how the brain functions. An original component of the materials is its focus on modeling choices, ie how do we choose the right approach, how do we build models, and how can we evaluate models to determine if they provide real (meaningful) insight. This meta-modeling component of the instructional materials asks what questions can be answered by different techniques, and how to apply them meaningfully to get insight about brain function.
@article{t2022neuromatch, doi = {10.21105/jose.00118}, title = {Neuromatch Academy: a 3-week, online summer school in computational neuroscience}, author = {t Hart, Bernard Marius and Achakulvisut, Titipat and Adeyemi, Ayoade and Akrami, Athena and Alicea, Bradly and Alonso-Andres, Alicia and Alzate-Correa, Diego and Ash, Arash and Ballesteros, Jesus J and Balwani, Aishwarya and others}, volume = {5}, number = {49}, pages = {118}, year = {2022}, journal = {Journal of Open Source Education} }
2021
- bioRxivDemuxalot: scaled up genetic demultiplexing for single-cell sequencingAlex Rogozhnikov, Pavan Ramkumar, Kevan Shah, and 3 more authorsbioRxiv, 2021
Demultiplexing methods have facilitated the widespread use of single-cell RNA sequencing (scRNAseq) experiments by lowering costs and reducing technical variations. Here, we present demuxalot: a method for probabilistic genotype inference from aligned reads, with no assumptions about allele ratios and efficient incorporation of prior genotype information from historical experiments in a multi-batch setting. Our method efficiently incorporates additional information across reads originating from the same transcript, enabling up to 3x more calls per read relative to naive approaches. We also propose a novel and highly performant tradeoff between methods that rely on reference genotypes and methods that learn variants from the data, by selecting a small number of highly informative variants that maximize the marginal information with respect to reference single nucleotide variants (SNVs). Our resulting improved SNV-based demultiplex method is up to 3x faster, 3x more data efficient, and achieves significantly more accurate doublet discrimination than previously published methods. This approach renders scRNAseq feasible for the kind of large multi-batch, multi-donor studies that are required to prosecute diseases with heterogeneous genetic backgrounds.
@article{rogozhnikov2021demuxalot, doi = {10.1101/2021.05.22.443646}, title = {Demuxalot: scaled up genetic demultiplexing for single-cell sequencing}, author = {Rogozhnikov, Alex and Ramkumar, Pavan and Shah, Kevan and Bedi, Rishi and Kato, Saul and Escola, G Sean}, journal = {bioRxiv}, pages = {2021--05}, year = {2021}, publisher = {Cold Spring Harbor Laboratory} } - Trends Cog SciNeuromatch Academy: Teaching computational neuroscience with global accessibilityTara Viegen, Athena Akrami, Kathryn Bonnen, and 8 more authorsTrends in Cognitive Sciences, 2021
Neuromatch Academy (NMA) designed and ran a fully online 3-week Computational Neuroscience Summer School for 1757 students with 191 teaching assistants (TAs) working in virtual inverted (or flipped) classrooms and on small group projects. Fourteen languages, active community management, and low cost allowed for an unprecedented level of inclusivity and universal accessibility.
@article{van2021neuromatch, doi = {}, title = {Neuromatch Academy: Teaching computational neuroscience with global accessibility}, author = {van Viegen, Tara and Akrami, Athena and Bonnen, Kathryn and DeWitt, Eric and Hyafil, Alexandre and Ledmyr, Helena and Lindsay, Grace W and Mineault, Patrick and Murray, John D and Pitkow, Xaq and others}, journal = {Trends in Cognitive Sciences}, volume = {25}, number = {7}, pages = {535--538}, year = {2021}, publisher = {Elsevier} } - Cell Reports
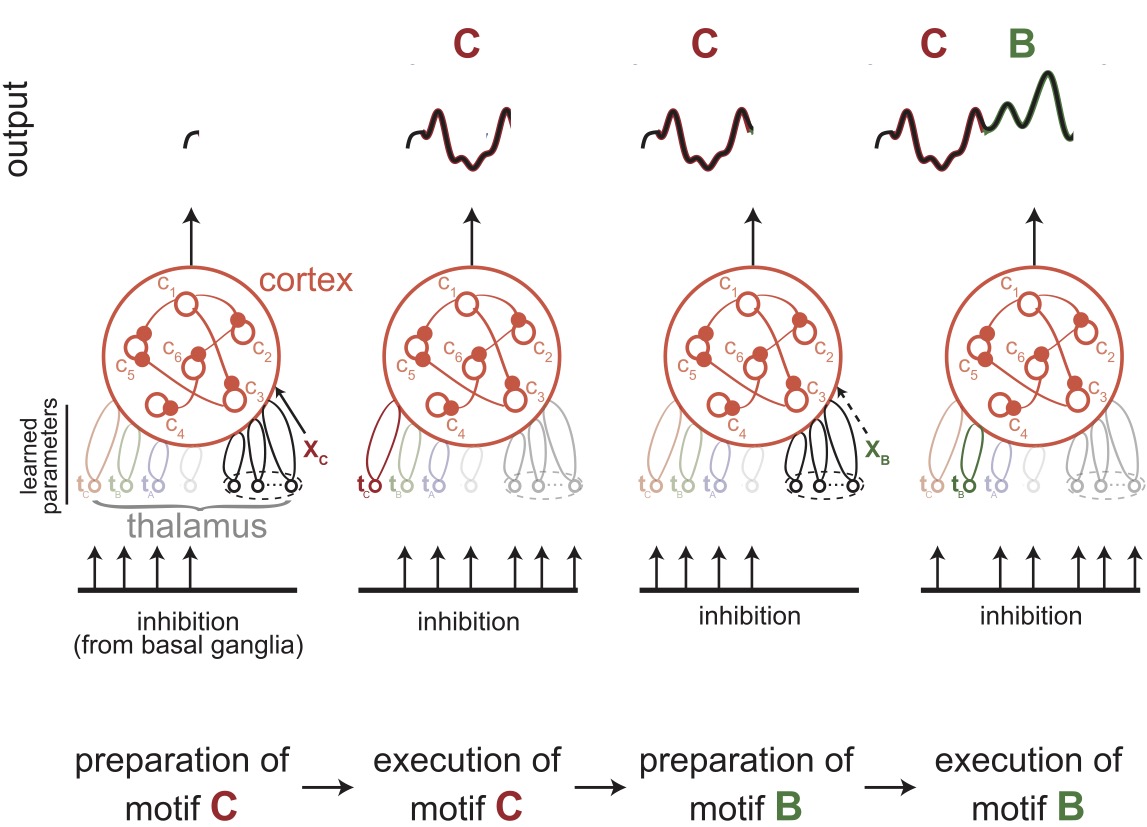 Thalamic control of cortical dynamics in a model of flexible motor sequencingLaureline Logiaco, LF Abbott, and Sean EscolaCell Reports, 2021
Thalamic control of cortical dynamics in a model of flexible motor sequencingLaureline Logiaco, LF Abbott, and Sean EscolaCell Reports, 2021The neural mechanisms that generate an extensible library of motor motifs and flexibly string them into arbitrary sequences are unclear. We developed a model in which inhibitory basal ganglia output neurons project to thalamic units that are themselves bidirectionally connected to a recurrent cortical network. We model the basal ganglia inhibitory patterns as silencing some thalamic neurons while leaving others disinhibited and free to interact with cortex during specific motifs. We show that a small number of disinhibited thalamic neurons can control cortical dynamics to generate specific motor output in a noise-robust way. Additionally, a single “preparatory” thalamocortical network can produce fast cortical dynamics that support rapid transitions between any pair of learned motifs. If the thalamic units associated with each sequence component are segregated, many motor outputs can be learned without interference and then combined in arbitrary orders for the flexible production of long and complex motor sequences.
@article{logiaco2021thalamic, doi = {10.1016/j.celrep.2021.109090}, title = {Thalamic control of cortical dynamics in a model of flexible motor sequencing}, author = {Logiaco, Laureline and Abbott, LF and Escola, Sean}, journal = {Cell Reports}, volume = {35}, number = {9}, year = {2021}, publisher = {Elsevier} }
2020
- bioRxivOptimization and scaling of patient-derived brain organoids uncovers deep phenotypes of diseaseKevan Shah, Rishi Bedi, Alex Rogozhnikov, and 8 more authorsbioRxiv, 2020
Cerebral organoids provide unparalleled access to human brain development in vitro. However, variability induced by current culture methodologies precludes using organoids as robust disease models. To address this, we developed an automated Organoid Culture and Assay (ORCA) system to support longitudinal unbiased phenotyping of organoids at scale across multiple patient lines. We then characterized organoid variability using novel machine learning methods and found that the contribution of donor, clone, and batch is significant and remarkably consistent over gene expression, morphology, and cell-type composition. Next, we performed multi-factorial protocol optimization, producing a directed forebrain protocol compatible with 96-well culture that exhibits low variability while preserving tissue complexity. Finally, we used ORCA to study tuberous sclerosis, a disease with known genetics but poorly representative animal models. For the first time, we report highly reproducible early morphological and molecular signatures of disease in heterozygous TSC+/− forebrain organoids, demonstrating the benefit of a scaled organoid system for phenotype discovery in human disease models.
@article{shah2020optimization, doi = {10.1101/2020.08.26.251611}, title = {Optimization and scaling of patient-derived brain organoids uncovers deep phenotypes of disease}, author = {Shah, Kevan and Bedi, Rishi and Rogozhnikov, Alex and Ramkumar, Pavan and Tong, Zhixiang and Rash, Brian and Stanton, Morgan and Sorokin, Jordan and Apaydin, Cagsar and Batarse, Anthony and others}, journal = {bioRxiv}, pages = {2020--08}, year = {2020}, publisher = {Cold Spring Harbor Laboratory} } - arXiv
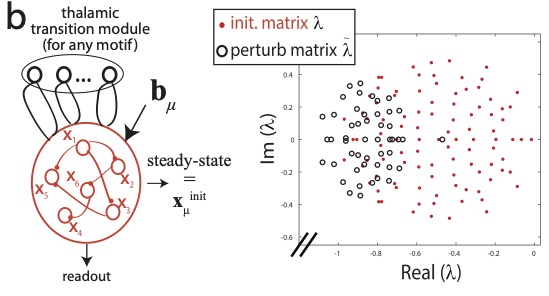 Thalamocortical motor circuit insights for more robust hierarchical control of complex sequencesLaureline Logiaco, and G Sean EscolaarXiv, 2020
Thalamocortical motor circuit insights for more robust hierarchical control of complex sequencesLaureline Logiaco, and G Sean EscolaarXiv, 2020We study learning of recurrent neural networks that produce temporal sequences consisting of the concatenation of re-usable "motifs". In the context of neuroscience or robotics, these motifs would be the motor primitives from which complex behavior is generated. Given a known set of motifs, can a new motif be learned without affecting the performance of the known set and then used in new sequences without first explicitly learning every possible transition? Two requirements enable this: (i) parameter updates while learning a new motif do not interfere with the parameters used for the previously acquired ones; and (ii) a new motif can be robustly generated when starting from the network state reached at the end of any of the other motifs, even if that state was not present during training. We meet the first requirement by investigating artificial neural networks (ANNs) with specific architectures, and attempt to meet the second by training them to generate motifs from random initial states. We find that learning of single motifs succeeds but that sequence generation is not robust: transition failures are observed. We then compare these results with a model whose architecture and analytically-tractable dynamics are inspired by the motor thalamocortical circuit, and that includes a specific module used to implement motif transitions. The synaptic weights of this model can be adjusted without requiring stochastic gradient descent (SGD) on the simulated network outputs, and we have asymptotic guarantees that transitions will not fail. Indeed, in simulations, we achieve single-motif accuracy on par with the previously studied ANNs and have improved sequencing robustness with no transition failures. Finally, we show that insights obtained by studying the transition subnetwork of this model can also improve the robustness of transitioning in the traditional ANNs previously studied.
@article{logiaco2020thalamocortical, doi = {10.48550/arXiv.2006.13332}, title = {Thalamocortical motor circuit insights for more robust hierarchical control of complex sequences}, author = {Logiaco, Laureline and Escola, G Sean}, journal = {arXiv}, year = {2020} } - Nat Comm
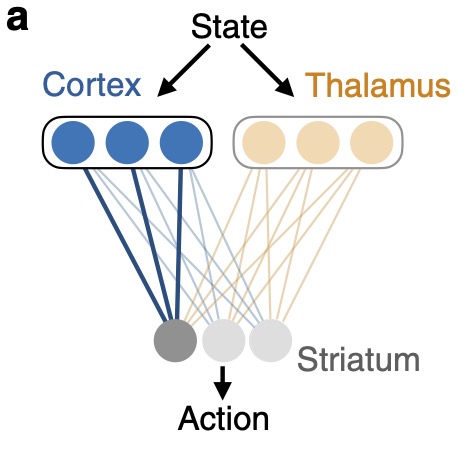 Remembrance of things practiced with fast and slow learning in cortical and subcortical pathwaysJames M Murray, and G Sean EscolaNature Communications, 2020
Remembrance of things practiced with fast and slow learning in cortical and subcortical pathwaysJames M Murray, and G Sean EscolaNature Communications, 2020The learning of motor skills unfolds over multiple timescales, with rapid initial gains in performance followed by a longer period in which the behavior becomes more refined, habitual, and automatized. While recent lesion and inactivation experiments have provided hints about how various brain areas might contribute to such learning, their precise roles and the neural mechanisms underlying them are not well understood. In this work, we propose neural- and circuit-level mechanisms by which motor cortex, thalamus, and striatum support motor learning. In this model, the combination of fast cortical learning and slow subcortical learning gives rise to a covert learning process through which control of behavior is gradually transferred from cortical to subcortical circuits, while protecting learned behaviors that are practiced repeatedly against overwriting by future learning. Together, these results point to a new computational role for thalamus in motor learning and, more broadly, provide a framework for understanding the neural basis of habit formation and the automatization of behavior through practice.
@article{murray2020remembrance, doi = {10.1038/s41467-020-19788-5}, title = {Remembrance of things practiced with fast and slow learning in cortical and subcortical pathways}, author = {Murray, James M and Escola, G Sean}, journal = {Nature Communications}, volume = {11}, number = {1}, pages = {6441}, year = {2020}, publisher = {Nature Publishing Group UK London} }
2018
- PLOS ONE
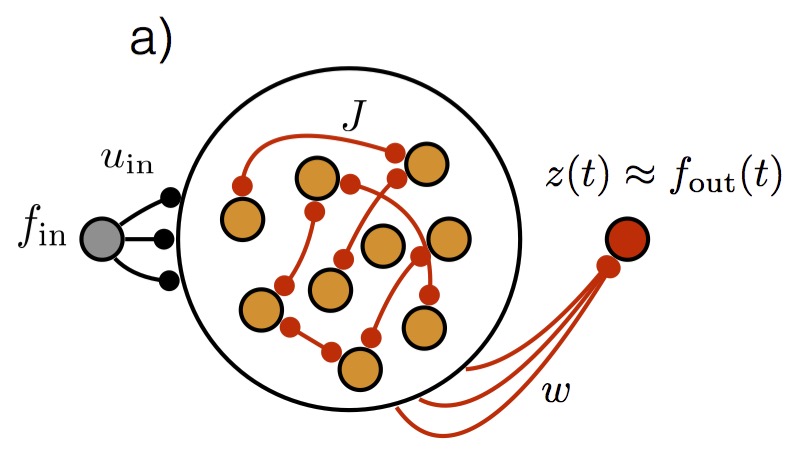 full-FORCE: A target-based method for training recurrent networksBrian DePasquale, Christopher J Cueva, Kanaka Rajan, and 2 more authorsPloS ONE, 2018
full-FORCE: A target-based method for training recurrent networksBrian DePasquale, Christopher J Cueva, Kanaka Rajan, and 2 more authorsPloS ONE, 2018Trained recurrent networks are powerful tools for modeling dynamic neural computations. We present a target-based method for modifying the full connectivity matrix of a recurrent network to train it to perform tasks involving temporally complex input/output transformations. The method introduces a second network during training to provide suitable “target” dynamics useful for performing the task. Because it exploits the full recurrent connectivity, the method produces networks that perform tasks with fewer neurons and greater noise robustness than traditional least-squares (FORCE) approaches. In addition, we show how introducing additional input signals into the target-generating network, which act as task hints, greatly extends the range of tasks that can be learned and provides control over the complexity and nature of the dynamics of the trained, task-performing network.
@article{depasquale2018full, doi = {10.1371/journal.pone.0191527}, title = {full-FORCE: A target-based method for training recurrent networks}, author = {DePasquale, Brian and Cueva, Christopher J and Rajan, Kanaka and Escola, G Sean and Abbott, LF}, journal = {PloS ONE}, volume = {13}, number = {2}, pages = {e0191527}, year = {2018}, publisher = {Public Library of Science San Francisco, CA USA} }
2017
- eLife
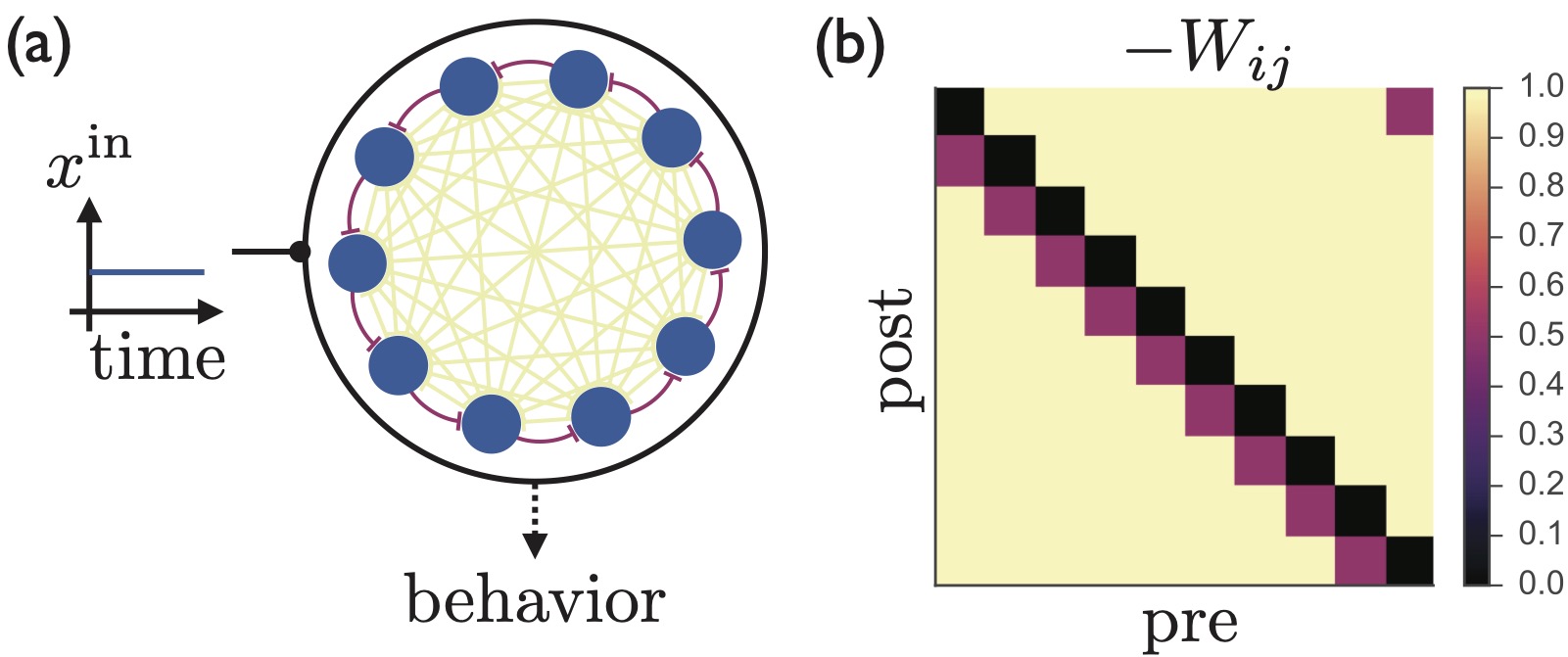 Learning multiple variable-speed sequences in striatum via cortical tutoringJames M Murray, and G Sean EscolaeLife, 2017
Learning multiple variable-speed sequences in striatum via cortical tutoringJames M Murray, and G Sean EscolaeLife, 2017Sparse, sequential patterns of neural activity have been observed in numerous brain areas during timekeeping and motor sequence tasks. Inspired by such observations, we construct a model of the striatum, an all-inhibitory circuit where sequential activity patterns are prominent, addressing the following key challenges: (i) obtaining control over temporal rescaling of the sequence speed, with the ability to generalize to new speeds; (ii) facilitating flexible expression of distinct sequences via selective activation, concatenation, and recycling of specific subsequences; and (iii) enabling the biologically plausible learning of sequences, consistent with the decoupling of learning and execution suggested by lesion studies showing that cortical circuits are necessary for learning, but that subcortical circuits are sufficient to drive learned behaviors. The same mechanisms that we describe can also be applied to circuits with both excitatory and inhibitory populations, and hence may underlie general features of sequential neural activity pattern generation in the brain.
@article{murray2017learning, doi = {10.7554/eLife.26084}, title = {Learning multiple variable-speed sequences in striatum via cortical tutoring}, author = {Murray, James M and Escola, G Sean}, journal = {eLife}, volume = {6}, pages = {e26084}, year = {2017}, publisher = {eLife Sciences Publications, Ltd} }
2011
- Neural Comp
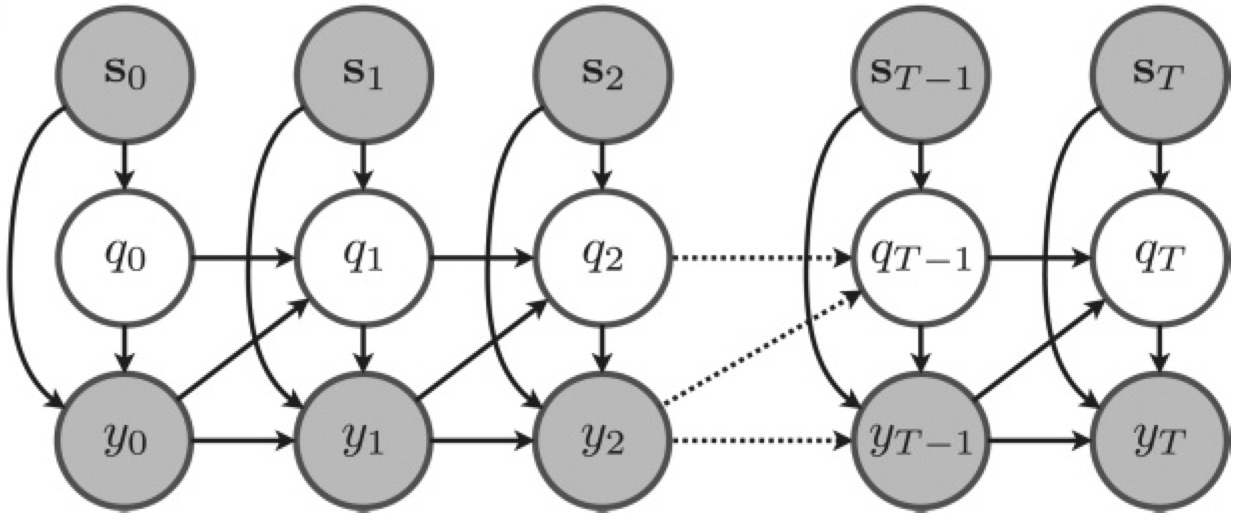 Hidden Markov models for the stimulus-response relationships of multistate neural systemsSean Escola, Alfredo Fontanini, Don Katz, and 1 more authorNeural Computation, 2011
Hidden Markov models for the stimulus-response relationships of multistate neural systemsSean Escola, Alfredo Fontanini, Don Katz, and 1 more authorNeural Computation, 2011Given recent experimental results suggesting that neural circuits may evolve through multiple firing states, we develop a framework for estimating state-dependent neural response properties from spike train data. We modify the traditional hidden Markov model (HMM) framework to incorporate stimulus-driven, non-Poisson point-process observations. For maximal flexibility, we allow external, time-varying stimuli and the neurons’ own spike histories to drive both the spiking behavior in each state and the transitioning behavior between states. We employ an appropriately modified expectation-maximization algorithm to estimate the model parameters. The expectation step is solved by the standard forward-backward algorithm for HMMs. The maximization step reduces to a set of separable concave optimization problems if the model is restricted slightly. We first test our algorithm on simulated data and are able to fully recover the parameters used to generate the data and accurately recapitulate the sequence of hidden states. We then apply our algorithm to a recently published data set in which the observed neuronal ensembles displayed multistate behavior and show that inclusion of spike history information significantly improves the fit of the model. Additionally, we show that a simple reformulation of the state space of the underlying Markov chain allows us to implement a hybrid half-multistate, half-histogram model that may be more appropriate for capturing the complexity of certain data sets than either a simple HMM or a simple peristimulus time histogram model alone.
@article{escola2011hidden, doi = {10.1162/NECO_a_00118}, title = {Hidden Markov models for the stimulus-response relationships of multistate neural systems}, author = {Escola, Sean and Fontanini, Alfredo and Katz, Don and Paninski, Liam}, journal = {Neural Computation}, volume = {23}, number = {5}, pages = {1071--1132}, year = {2011}, publisher = {MIT Press} }
2009
- Markov chains, neural responses, and optimal temporal computationsGary Sean Escola2009
@book{escola2009markov, title = {Markov chains, neural responses, and optimal temporal computations}, author = {Escola, Gary Sean}, year = {2009}, publisher = {Columbia University} } - Neural Comp
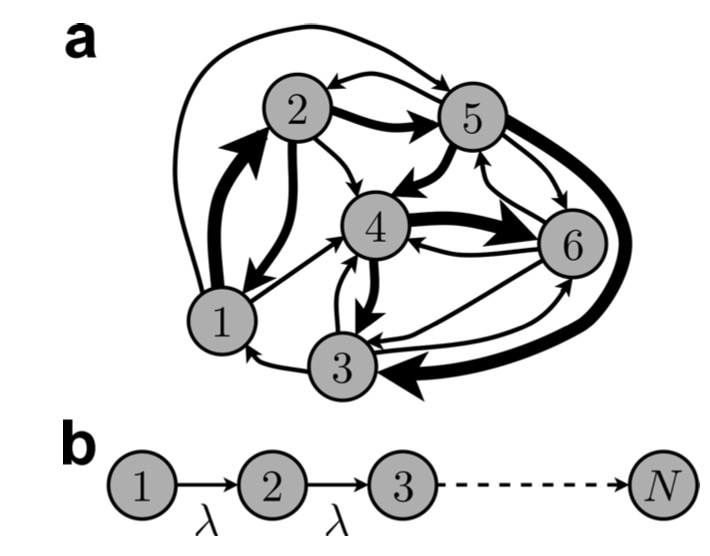 Maximally reliable Markov chains under energy constraintsSean Escola, Michael Eisele, Kenneth Miller, and 1 more authorNeural Computation, 2009
Maximally reliable Markov chains under energy constraintsSean Escola, Michael Eisele, Kenneth Miller, and 1 more authorNeural Computation, 2009Signal-to-noise ratios in physical systems can be significantly degraded if the outputs of the systems are highly variable. Biological processes for which highly stereotyped signal generations are necessary features appear to have reduced their signal variabilities by employing multiple processing steps. To better understand why this multistep cascade structure might be desirable, we prove that the reliability of a signal generated by a multistate system with no memory (i.e., a Markov chain) is maximal if and only if the system topology is such that the process steps irreversibly through each state, with transition rates chosen such that an equal fraction of the total signal is generated in each state. Furthermore, our result indicates that by increasing the number of states, it is possible to arbitrarily increase the reliability of the system. In a physical system, however, an energy cost is associated with maintaining irreversible transitions, and this cost increases with the number of such transitions (i.e., the number of states). Thus, an infinite-length chain, which would be perfectly reliable, is infeasible. To model the effects of energy demands on the maximally reliable solution, we numerically optimize the topology under two distinct energy functions that penalize either irreversible transitions or incommunicability between states, respectively. In both cases, the solutions are essentially irreversible linear chains, but with upper bounds on the number of states set by the amount of available energy. We therefore conclude that a physical system for which signal reliability is important should employ a linear architecture, with the number of states (and thus the reliability) determined by the intrinsic energy constraints of the system.
@article{escola2009maximally, doi = {10.1162/neco.2009.08-08-843}, title = {Maximally reliable Markov chains under energy constraints}, author = {Escola, Sean and Eisele, Michael and Miller, Kenneth and Paninski, Liam}, journal = {Neural Computation}, volume = {21}, number = {7}, pages = {1863--1912}, year = {2009}, publisher = {MIT Press One Rogers Street, Cambridge, MA 02142-1209, USA journals-info~…} }
2007
- Annals NYASReconstruction of metabolic networks from high-throughput metabolite profiling data: in silico analysis of red blood cell metabolismIlya Nemenman, G Sean Escola, William S Hlavacek, and 3 more authorsAnnals of the New York Academy of Sciences, 2007
We investigate the ability of algorithms developed for reverse engineering of transcriptional regulatory networks to reconstruct metabolic networks from high-throughput metabolite profiling data. For benchmarking purposes, we generate synthetic metabolic profiles based on a well-established model for red blood cell metabolism. A variety of data sets are generated, accounting for different properties of real metabolic networks, such as experimental noise, metabolite correlations, and temporal dynamics. These data sets are made available online. We use ARACNE, a mainstream algorithm for reverse engineering of transcriptional regulatory networks from gene expression data, to predict metabolic interactions from these data sets. We find that the performance of ARACNE on metabolic data is comparable to that on gene expression data.
@article{nemenman2007reconstruction, doi = {10.1196/annals.1407.013}, title = {Reconstruction of metabolic networks from high-throughput metabolite profiling data: in silico analysis of red blood cell metabolism}, author = {Nemenman, Ilya and Escola, G Sean and Hlavacek, William S and Unkefer, Pat J and Unkefer, Clifford J and Wall, Michael E}, journal = {Annals of the New York Academy of Sciences}, volume = {1115}, number = {1}, pages = {102--115}, year = {2007}, publisher = {Wiley Online Library} }